Monitor the training process
Overview
Teaching: 135 min
Exercises: 80 minQuestions
How do I set the training goal?
How do I monitor the training process?
How do I detect (and avoid) overfitting?
What are common options to improve the model performance?
Objectives
Explain the importance of splitting the training data
Use the data splits to plot the training process
Set the training goal for your deep neural network
Measure the performance of your deep neural network
Interpret the training plots to recognize overfitting
Implement basic strategies to prevent overfitting
Import & explore the data
Import dataset
Here we want to work with the weather prediction dataset (the light version) which can be
downloaded from Zenodo.
It contains daily weather observations from 11 different European cities or places through the
years 2000 to 2010. For all locations the data contains the variables ‘mean temperature’, ‘max temperature’, and ‘min temperature’. In addition, for multiple of the following variables are provided: ‘cloud_cover’, ‘wind_speed’, ‘wind_gust’, ‘humidity’, ‘pressure’, ‘global_radiation’, ‘precipitation’, ‘sunshine’, but not all of them are provided for all locations. A more extensive description of the dataset including the different physical units is given in accompanying metadata file.
import pandas as pd
filename_data = "weather_prediction_dataset_light.csv"
data = pd.read_csv(filename_data)
data.head()
| DATE | MONTH | BASEL_cloud_cover | BASEL_humidity | BASEL_pressure | … | |
|---|---|---|---|---|---|---|
| 0 | 20000101 | 1 | 8 | 0.89 | 1.0286 | … |
| 1 | 20000102 | 1 | 8 | 0.87 | 1.0318 | … |
| 2 | 20000103 | 1 | 5 | 0.81 | 1.0314 | … |
| 3 | 20000104 | 1 | 7 | 0.79 | 1.0262 | … |
| 4 | 20000105 | 1 | 5 | 0.90 | 1.0246 | … |
Load the data
If you have not downloaded the data yet, you can also load it directly from Zenodo:
data = pd.read_csv("https://zenodo.org/record/5071376/files/weather_prediction_dataset_light.csv?download=1")
Brief exploration of the data
Let us start with a quick look at the type of features that we find in the data.
data.columns
Index(['DATE', 'MONTH', 'BASEL_cloud_cover', 'BASEL_humidity',
'BASEL_pressure', 'BASEL_global_radiation', 'BASEL_precipitation',
'BASEL_sunshine', 'BASEL_temp_mean', 'BASEL_temp_min', 'BASEL_temp_max',
...
'SONNBLICK_temp_min', 'SONNBLICK_temp_max', 'TOURS_humidity',
'TOURS_pressure', 'TOURS_global_radiation', 'TOURS_precipitation',
'TOURS_temp_mean', 'TOURS_temp_min', 'TOURS_temp_max'],
dtype='object')
Exercise: Explore the dataset
Let’s get a quick idea of the dataset.
- How many data points do we have?
- How many features does the data have (don’t count month and date as a feature)?
- What are the different measured variable types in the data and how many are there (humidity etc.) ?
Solution
data.shapeThis will give both the number of datapoints (3654) and the number of features (89 + month + date).
To see what type of features the data contains we could run something like:
print({x.split("_")[-1] for x in data.columns if x not in ["MONTH", "DATE"]}){'precipitation', 'max', 'radiation', 'humidity', 'sunshine', 'min', 'pressure', 'mean', 'cover'}An alternative way which is slightly more complicated but gives better results is using regex.
import re feature_names = set() for col in data.columns: feature_names.update(re.findall('[^A-Z]{2,}', col)) feature_namesIn total there are 9 different measured variables.
Define the problem: Predict tomorrow’s sunshine hours
Select a subset and split into data (X) and labels (y)
The full dataset comprises 10 years (3654 days) from which we here will only select the first 3 years.
We will then define what exactly we want to predict from this data. A very common task with weather data is to make a prediction about the weather sometime in the future, say the next day. The present dataset is sorted by “DATE”, so for each row i in the table we can pick a corresponding feature and location from row i+1 that we later want to predict with our model.
Here we will pick a rather difficult-to-predict feature, sunshine hours, which we want to predict for the location: BASEL.
nr_rows = 365*3
# data
X_data = data.loc[:nr_rows].drop(columns=['DATE', 'MONTH'])
# labels (sunshine hours the next day)
y_data = data.loc[1:(nr_rows + 1)]["BASEL_sunshine"]
Prepare the data for machine learning
In general, it is important to check if the data contains any unexpected values such as 9999 or NaN or NoneType. You can use the pandas data.describe() function for this. If so, such values must be removed or replaced.
In the present case the data is luckily well prepared and shouldn’t contain such values, so that this step can be omitted.
Split data and labels into training, validation, and test set
As with classical machine learning techniques, it is required in deep learning to split off a hold-out test set which remains untouched during model training and tuning. It is later used to evaluate the model performance. On top, we will also split off an additional validation set, the reason of which will hopefully become clearer later in this lesson.
To make our lives a bit easier, we employ a trick to create these 3 datasets, training set, test set and validation set, by calling the train_test_split method of scikit-learn twice.
First we create the training set and leave the remainder of 30 % of the data to the two hold-out sets.
from sklearn.model_selection import train_test_split
X_train, X_holdout, y_train, y_holdout = train_test_split(X_data, y_data, test_size=0.3, random_state=0)
Now we split the 30 % of the data in two equal sized parts.
X_val, X_test, y_val, y_test = train_test_split(X_holdout, y_holdout, test_size=0.5, random_state=0)
Setting the random_state to 0 is a short-hand at this point. Note however, that changing this seed of the pseudo-random number generator will also change the composition of your data sets. For the sake of reproducibility, this is one example of a parameters that should not change at all.
Build a dense neural network
Regression and classification - how to set a training goal
In episode 2 we trained a dense neural network on a classification task. For this one hot encoding was used together with a Categorical Crossentropy loss function.
This measured how close the distribution of the neural network outputs corresponds to the distribution of the three values in the one hot encoding.
Now we want to work on a regression task, thus not predicting a class label (or integer number) for a datapoint. In regression, we like to predict one (and sometimes many) values of a feature. This is typically a floating point number.
Exercise: Architecture of the network
As we want to design a neural network architecture for a regression task, see if you can first come up with the answers to the following questions:
- What must be the dimension of our input layer?
- We want to to output the prediction of a single number. The output layer of the NN hence cannot be the same as for the classification task earlier. This is because the
softmaxactivation being used had a concrete meaning with respect to the class labels which is not needed here. What output layer design would you choose for regression?
Hint: A layer withreluactivation, withsigmoidactivation or no activation at all?Solution
- The shape of the input layer has to correspond to the number of features in our data: 89
- The output is a single value per prediction, so the output layer can consist of a dense layer with only one node. The softmax activiation function works well for a classification task, but here we do not want to restrict the possible outcomes to the range of zero and one. In fact, we can omit the activation in the output layer.
In our example we want to predict the sunshine hours in Basel (or any other place in the dataset) for tomorrow based on the weather data of all 18 locations today. BASEL_sunshine is a floating point value (i.e. float64). The network should hence output a single float value which is why the last layer of our network will only consist of a single node.
We compose a network of two hidden layers to start off with something. We go by a scheme with 100 neurons in the first hidden layer and 50 neurons in the second layer. As activation function we settle on the relu function as a it proved very robust and widely used. To make our live easier later, we wrap the definition of the network in a method called create_nn.
from tensorflow import keras
def create_nn():
# Input layer
inputs = keras.Input(shape=(X_data.shape[1],), name='input')
# Dense layers
layers_dense = keras.layers.Dense(100, 'relu')(inputs)
layers_dense = keras.layers.Dense(50, 'relu')(layers_dense)
# Output layer
outputs = keras.layers.Dense(1)(layers_dense)
return keras.Model(inputs=inputs, outputs=outputs, name="weather_prediction_model")
model = create_nn()
The shape of the input layer has to correspond to the number of features in our data: 89. We use X_data.shape[1] to obtain this value dynamically
The output layer here is a dense layer with only 1 node. And we here have chosen to use no activation function. While we might use softmax for a classification task, here we do not want to restrict the possible outcomes for a start.
In addition, we have here chosen to write the network creation as a function so that we can use it later again to initiate new models.
Let’s check how our model looks like by calling the summary method.
model.summary()
Model: "weather_prediction_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input (InputLayer) [(None, 89)] 0
_________________________________________________________________
dense (Dense) (None, 100) 9000
_________________________________________________________________
dense_1 (Dense) (None, 50) 5050
_________________________________________________________________
dense_2 (Dense) (None, 1) 51
=================================================================
Total params: 14,101
Trainable params: 14,101
Non-trainable params: 0
When compiling the model we can define a few very important aspects. We will discuss them now in more detail.
Loss function
The loss is what the neural network will be optimized on during training, so choosing a suitable loss function is crucial for training neural networks.
In the given case we want to stimulate that the predicted values are as close as possible to the true values. This is commonly done by using the mean squared error (mse) or the mean absolute error (mae), both of which should work OK in this case. Often, mse is preferred over mae because it “punishes” large prediction errors more severely.
In Keras this is implemented in the keras.losses.MeanSquaredError class (see Keras documentation: https://keras.io/api/losses/). This can be provided into the model.compile method with the loss parameter and setting it to mse, e.g.
model.compile(loss='mse')
Optimizer
Somewhat coupled to the loss function is the optimizer that we want to use. The optimizer here refers to the algorithm with which the model learns to optimize on the provided loss function. A basic example for such an optimizer would be stochastic gradient descent. For now, we can largely skip this step and pick one of the most common optimizers that works well for most tasks: the Adam optimizer. Similar to activation functions, the choice of optimizer depends on the problem you are trying to solve, your model architecture and your data. Adam is a good starting point though, which is why we chose it.
model.compile(optimizer='adam',
loss='mse')
Metrics
In our first example (episode 2) we plotted the progression of the loss during training.
That is indeed a good first indicator if things are working alright, i.e. if the loss is indeed decreasing as it should with the number of epochs.
However, when models become more complicated then also the loss functions often become less intuitive.
That is why it is good practice to monitor the training process with additional, more intuitive metrics.
They are not used to optimize the model, but are simply recorded during training.
With Keras such additional metrics can be added via metrics=[...] parameter and can contain one or multiple metrics of interest.
Here we could for instance chose to use 'mae' the mean absolute error, or the the root mean squared error (RMSE) which unlike the mse has the same units as the predicted values. For the sake of units, we choose the latter.
model.compile(optimizer='adam',
loss='mse',
metrics=[keras.metrics.RootMeanSquaredError()])
With this, we complete the compilation of our network and are ready to start training.
Challenge: Metrics
Look into the Keras documentation on metrics. Choose an additional metric that you would like to track, and compile the model again with this metric added.
Solution
As we are facing a regression task, we should pick one of the metrics under Regression metrics. For example, if we choose Mean Absolute Error, we can compile the model as follows:
model.compile(optimizer='adam', loss='mse', metrics=[keras.metrics.RootMeanSquaredError(), keras.metrics.MeanAbsoluteError()])
Train a dense neural network
Now that we created and compiled our dense neural network, we can start training it.
One additional concept we need to introduce though, is the batch_size.
This defines how many samples from the training data will be used to estimate the error gradient before the model weights are updated.
Larger batches will produce better, more accurate gradient estimates but also less frequent updates of the weights.
Here we are going to use a batch size of 32 which is a common starting point.
history = model.fit(X_train, y_train,
batch_size=32,
epochs=200,
verbose=2)
We can plot the training process using the history object returned from the model training:
import seaborn as sns
import matplotlib.pyplot as plt
history_df = pd.DataFrame.from_dict(history.history)
sns.lineplot(data=history_df['root_mean_squared_error'])
plt.xlabel("epochs")
plt.ylabel("RMSE")
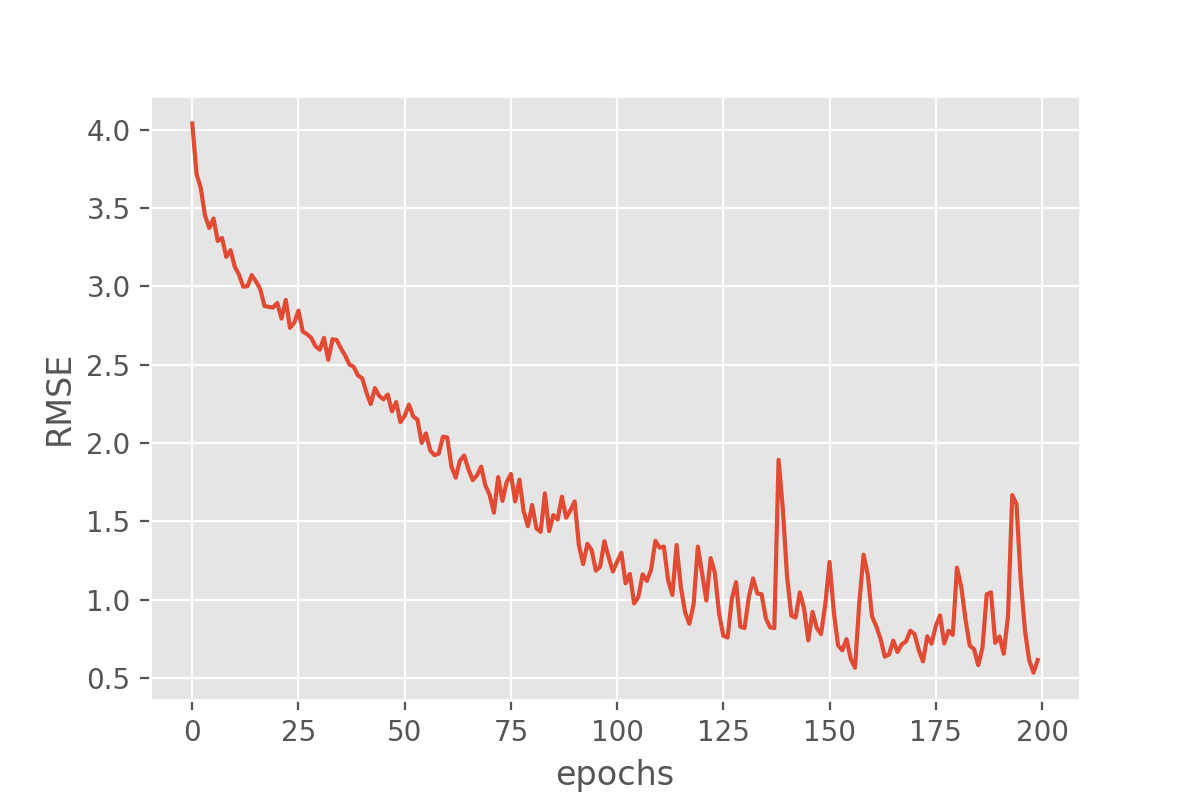
This looks very promising! Our metric (“RMSE”) is dropping nicely and while it maybe keeps fluctuating a bit it does end up at fairly low RMSE values. But the RMSE is just the root mean squared error, so we might want to look a bit more in detail how well our just trained model does in predicting the sunshine hours.
Evaluate our model
There is not a single way to evaluate how a model performs. But there are at least two very common approaches. For a classification task that is to compute a confusion matrix for the test set which shows how often particular classes were predicted correctly or incorrectly.
For the present regression task, it makes more sense to compare true and predicted values in a scatter plot.
First, we will do the actual prediction step.
y_train_predicted = model.predict(X_train)
y_test_predicted = model.predict(X_test)
So, let’s look at how the predicted sunshine hour have developed with reference to their ground truth values.
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
plt.style.use('ggplot') # optional, that's only to define a visual style
axes[0].scatter(y_train_predicted, y_train, s=10, alpha=0.5, color="teal")
axes[0].set_title("training set")
axes[0].set_xlabel("predicted sunshine hours")
axes[0].set_ylabel("true sunshine hours")
axes[1].scatter(y_test_predicted, y_test, s=10, alpha=0.5, color="teal")
axes[1].set_title("test set")
axes[1].set_xlabel("predicted sunshine hours")
axes[1].set_ylabel("true sunshine hours")
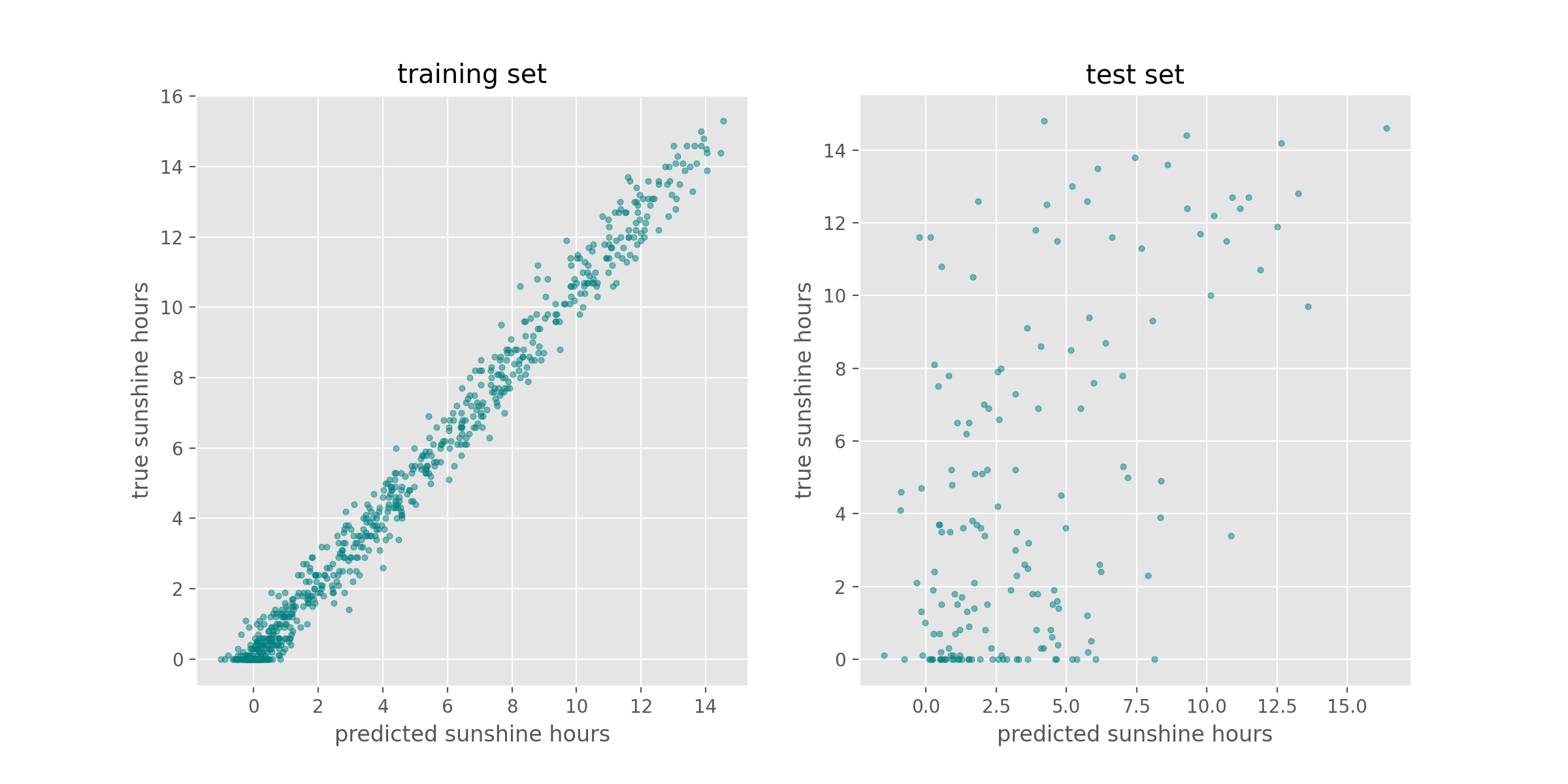
Exercise: Reflecting on our results
- Is the performance of the model as you expected (or better/worse)?
- Is there a noteable difference between training set and test set? And if so, any idea why?
Solution
While the performance on the train set seems reasonable, the performance on the test set is much worse. This is a common problem called overfitting, which we will discuss in more detail later.
The accuracy on the training set seems fairly good. In fact, considering that the task of predicting the daily sunshine hours is really not easy it might even be surprising how well the model predicts that (at least on the training set). Maybe a little too good? We also see the noticeable difference between train and test set when calculating the exact value of the RMSE:
train_metrics = model.evaluate(X_train, y_train, return_dict=True)
test_metrics = model.evaluate(X_test, y_test, return_dict=True)
print('Train RMSE: {:.2f}, Test RMSE: {:.2f}'.format(train_metrics['root_mean_squared_error'], test_metrics['root_mean_squared_error']))
24/24 [==============================] - 0s 442us/step - loss: 0.7092 - root_mean_squared_error: 0.8421
6/6 [==============================] - 0s 647us/step - loss: 16.4413 - root_mean_squared_error: 4.0548
Train RMSE: 0.84, Test RMSE: 4.05
For those experienced with (classical) machine learning this might look familiar. The plots above expose the signs of overfitting which means that the model has to some extent memorized aspects of the training data. As a result, it makes much more accurate predictions on the training data than on unseen test data.
Overfitting also happens in classical machine learning, but there it is usually interpreted as the model having more parameters than the training data would justify (say, a decision tree with too many branches for the number of training instances). As a consequence one would reduce the number of parameters to avoid overfitting. In deep learning the situation is slightly different. It can - as for classical machine learning - also be a sign of having a too big model, meaning a model with too many parameters (layers and/or nodes). However, in deep learning higher number of model parameters are often still considered acceptable and models often perform best (in terms of prediction accuracy) when they are at the verge of overfitting. So, in a way, training deep learning models is always a bit like playing with fire…
Set expectations: How difficult is the defined problem?
Before we dive deeper into handling overfitting and (trying to) improving the model performance, let’s ask the question: How well must a model perform before we consider it a good model?
Now that we defined a problem (predict tomorrow’s sunshine hours), it makes sense to develop an intuition for how difficult the posed problem is. Frequently, models will be evaluated against a so called baseline. A baseline can be the current standard in the field or if such a thing does not exist it could also be an intuitive first guess or toy model. The latter is exactly what we would use for our case.
Maybe the simplest sunshine hour prediction we can easily do is: Tomorrow we will have the same number of sunshine hours as today. (sounds very naive, but for many observables such as temperature this is already a fairly good predictor)
We can take the BASEL_sunshine column of our data, because this contains the sunshine hours from one day before what we have as a label.
y_baseline_prediction = X_test['BASEL_sunshine']
plt.figure(figsize=(5, 5), dpi=100)
plt.scatter(y_baseline_prediction, y_test, s=10, alpha=0.5)
plt.xlabel("sunshine hours yesterday")
plt.ylabel("true sunshine hours")
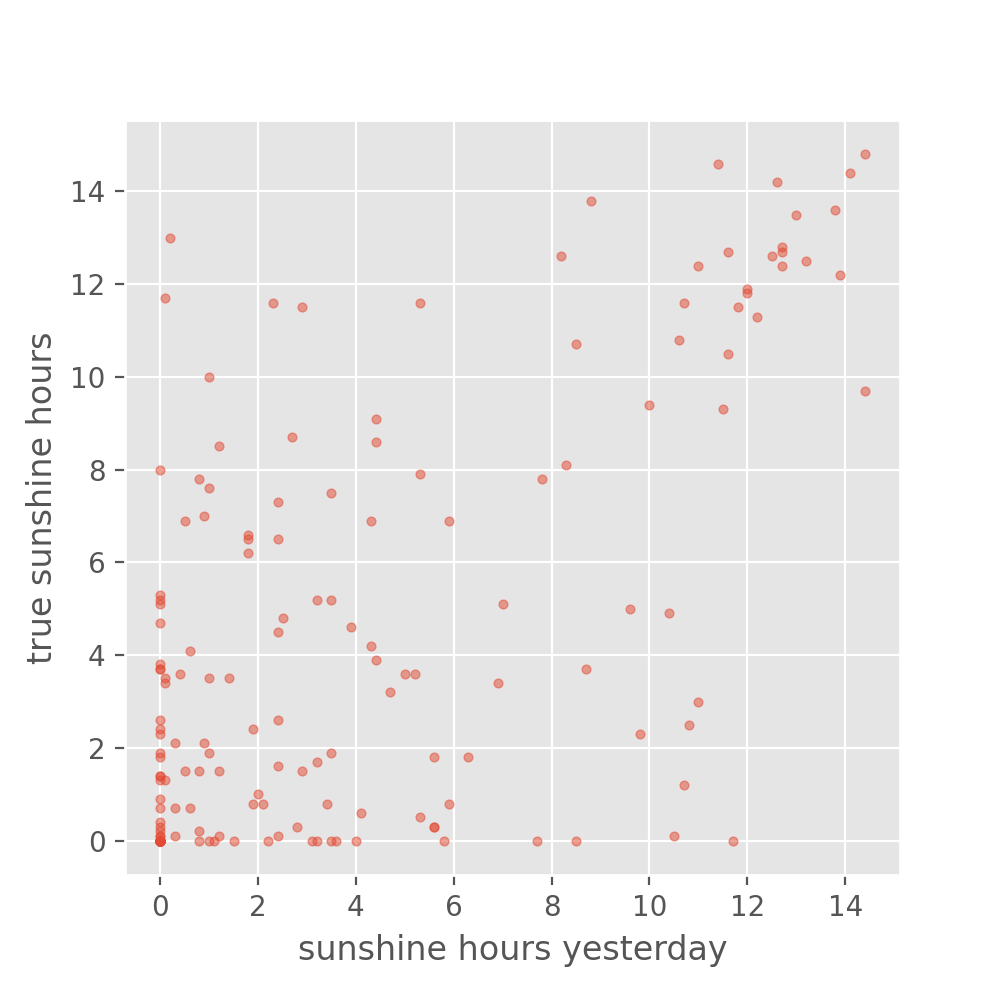
It is difficult to interpret from this plot whether our model is doing better than the baseline. We can also have a look at the RMSE:
from sklearn.metrics import mean_squared_error
rmse_nn = mean_squared_error(y_test, y_test_predicted, squared=False)
rmse_baseline = mean_squared_error(y_test, y_baseline_prediction, squared=False)
print('NN RMSE: {:.2f}, baseline RMSE: {:.2f}'.format(rmse_nn, rmse_baseline))
NN RMSE: 4.05, baseline RMSE: 3.88
Judging from the numbers alone, our neural network preduction would be performing worse than the baseline.
Exercise: Baseline
Looking at this baseline: Would you consider this a simple or a hard problem to solve?
Solution
This really depends on your definition of hard! The baseline gives a more accurate prediction than just randomly predicting a number, so the problem is not impossible to solve with machine learning. However, given the structure of the data and our expectations with respect to quality of prediction, it may remain hard to find a good algorithm which exceeds our baseline by orders of magnitude.
Watch your model training closely
As we saw when comparing the predictions for the training and the test set, deep learning models are prone to overfitting. Instead of iterating through countless cycles of model trainings and subsequent evaluations with a reserved test set, it is common practice to work with a second split off dataset to monitor the model during training. This is the validation set which can be regarded as a second test set. As with the test set, the datapoints of the validation set are not used for the actual model training itself. Instead, we evaluate the model with the validation set after every epoch during training, for instance to stop if we see signs of clear overfitting. Since we are adapting our model (tuning our hyperparameters) based on this validation set, it is very important that it is kept separate from the test set. If we used the same set, we wouldn’t know whether our model truly generalizes or is only overfitting.
Training vs. validation set
Not everybody agrees on the terminology of training set versus validation set. You might find examples in literature where these terms are used the other way around.
We are sticking to the definition that is consistent with the Keras API. In there, the validation set can be used during training, and the test set is reserved for afterwards.
Let’s give this a try!
We need to initiate a new model – otherwise Keras will simply assume that we want to continue training the model we already trained above.
model = create_nn()
model.compile(optimizer='adam',
loss='mse',
metrics=[keras.metrics.RootMeanSquaredError()])
But now we train it with the small addition of also passing it our validation set:
history = model.fit(X_train, y_train,
batch_size=32,
epochs=200,
validation_data=(X_val, y_val),
verbose=2)
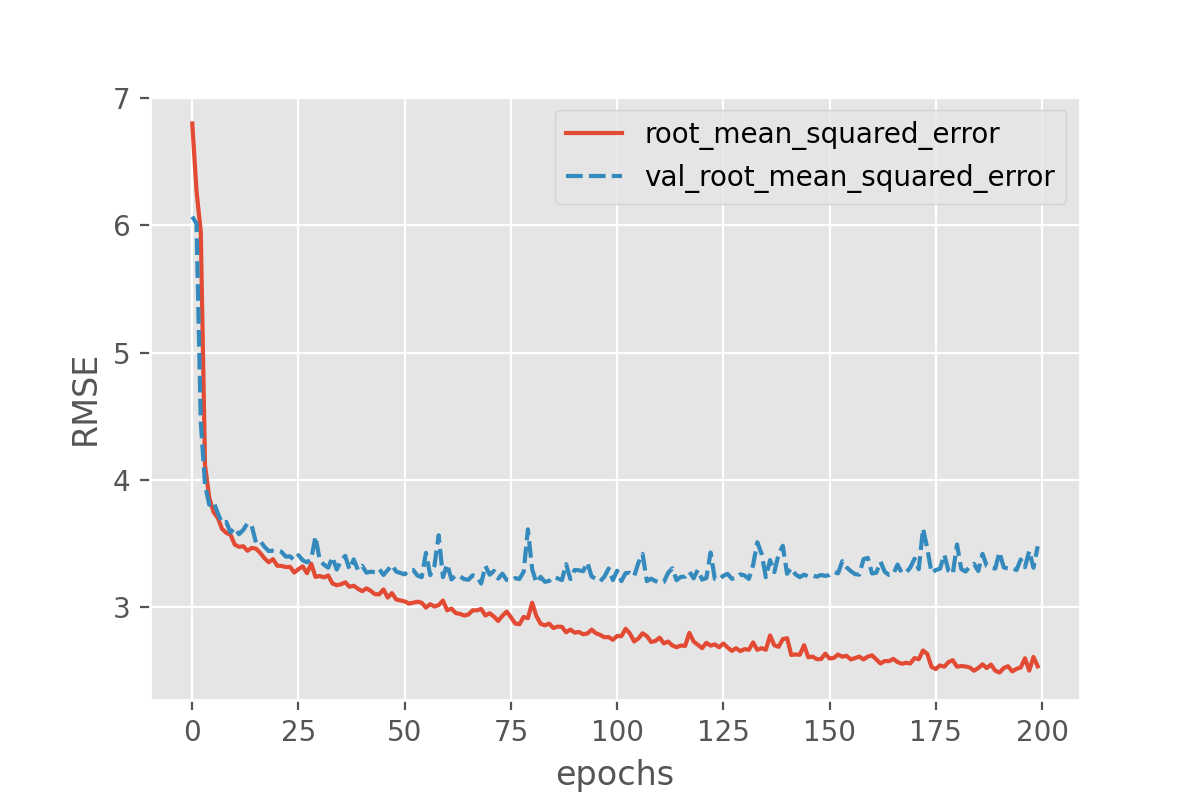
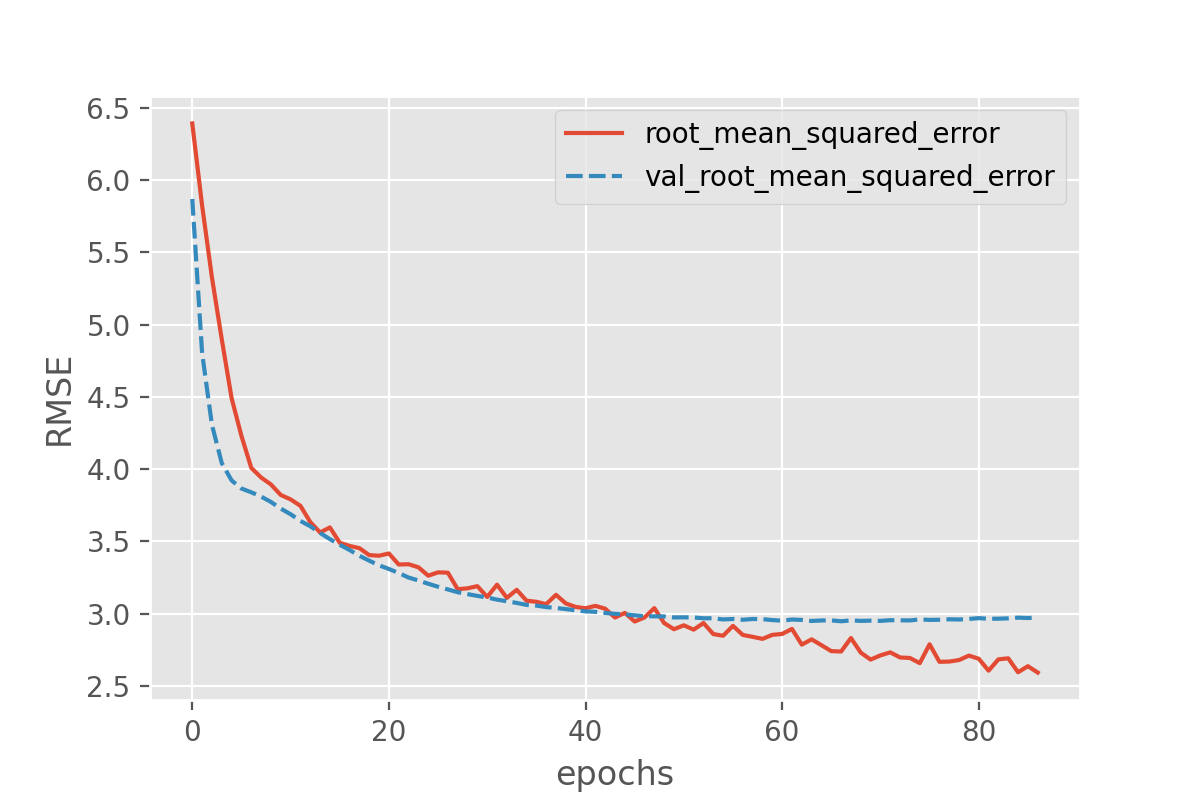
With this we can plot both the performance on the training data and on the validation data!
history_df = pd.DataFrame.from_dict(history.history)
sns.lineplot(data=history_df[['root_mean_squared_error', 'val_root_mean_squared_error']])
plt.xlabel("epochs")
plt.ylabel("RMSE")
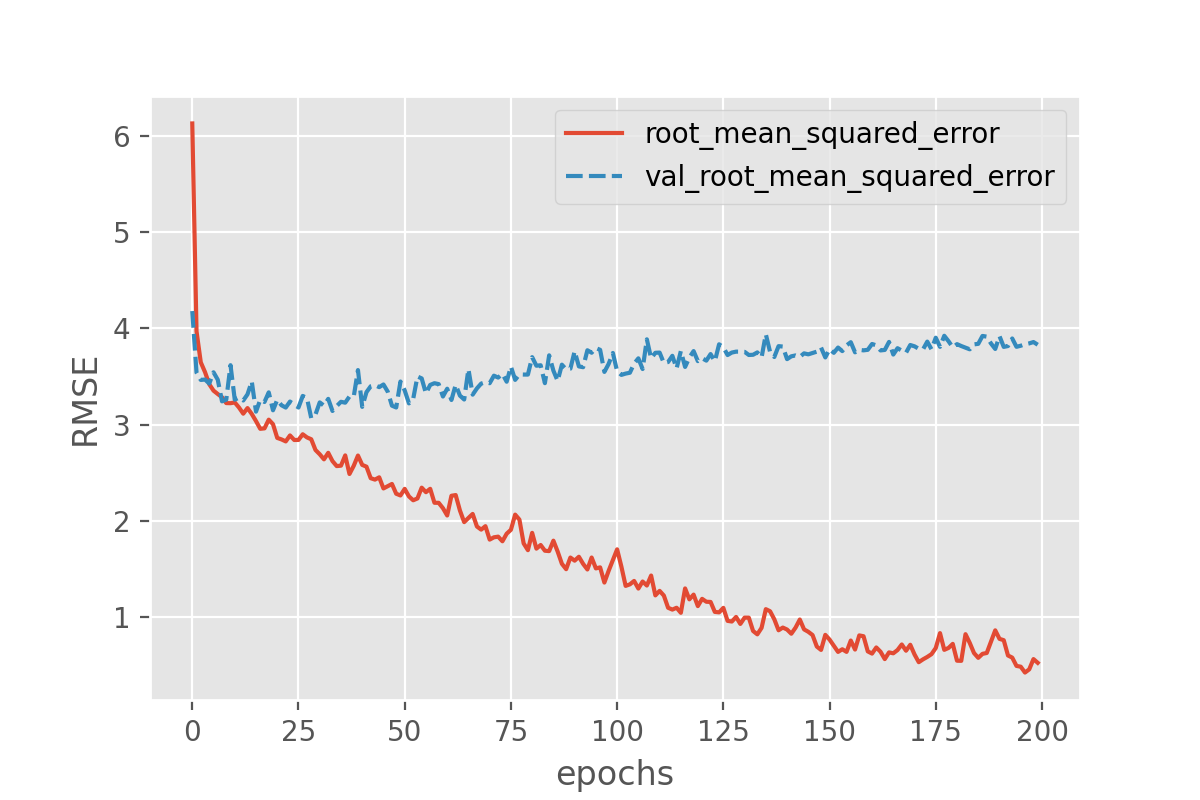
Exercise: plot the training progress.
Is there a difference between the training and validation data? And if so, what would this imply?
Solution
The difference between training and validation data shows that something is not completely right here. The model predictions on the validation set quickly seem to reach a plateau while the performance on the training set keeps improving. That is a common signature of overfitting.
Counteract model overfitting
Overfitting is a very common issue and there are many strategies to handle it. Most similar to classical machine learning might to reduce the number of parameters.
Try to reduce the degree of overfitting by lowering the number of parameters
We can keep the network architecture unchanged (2 dense layers + a one-node output layer) and only play with the number of nodes per layer. Try to lower the number of nodes in one or both of the two dense layers and observe the changes to the training and validation losses. If time is short: Suggestion is to run one network with only 10 and 5 nodes in the first and second layer.
- Is it possible to get rid of overfitting this way?
- Does the overall performance suffer or does it mostly stay the same?
- How low can you go with the number of parameters without notable effect on the performance on the validation set?
Solution
def create_nn(nodes1=100, nodes2=50): # Input layer inputs = keras.layers.Input(shape=(X_data.shape[1],), name='input') # Dense layers layers_dense = keras.layers.Dense(nodes1, 'relu')(inputs) layers_dense = keras.layers.Dense(nodes2, 'relu')(layers_dense) # Output layer outputs = keras.layers.Dense(1)(layers_dense) return keras.Model(inputs=inputs, outputs=outputs, name="model_small") model = create_nn(10, 5) model.summary()Model: "model_small" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= input (InputLayer) [(None, 89)] 0 _________________________________________________________________ dense_9 (Dense) (None, 10) 900 _________________________________________________________________ dense_10 (Dense) (None, 5) 55 _________________________________________________________________ dense_11 (Dense) (None, 1) 6 ================================================================= Total params: 961 Trainable params: 961 Non-trainable params: 0model.compile(optimizer='adam', loss='mse', metrics=[keras.metrics.RootMeanSquaredError()]) history = model.fit(X_train, y_train, batch_size = 32, epochs = 200, validation_data=(X_val, y_val), verbose = 2) history_df = pd.DataFrame.from_dict(history.history) sns.lineplot(data=history_df[['root_mean_squared_error', 'val_root_mean_squared_error']]) plt.xlabel("epochs") plt.ylabel("RMSE")
There is no single correct solution here. But you will have noticed that the number of nodes can be reduced quite a bit!
In general, it quickly becomes a very complicated search for the right “sweet spot”, i.e. the settings for which overfitting will be (nearly) avoided but which still performes equally well.
We saw that reducing the number of parameters can be a strategy to avoid overfitting. In practice, however, this is usually not the (main) way to go when it comes to deep learning. One reason is, that finding the sweet spot can be really hard and time consuming. And it has to be repeated every time the model is adapted, e.g. when more training data becomes available.
Sweet Spots
Note: There is no single correct solution here. But you will have noticed that the number of nodes can be reduced quite a bit! In general, it quickly becomes a very complicated search for the right “sweet spot”, i.e. the settings for which overfitting will be (nearly) avoided but which still performes equally well.
Early stopping: stop when things are looking best
Arguable the most common technique to avoid (severe) overfitting in deep learning is called early stopping. As the name suggests, this technique just means that you stop the model training if things do not seem to improve anymore. More specifically, this usually means that the training is stopped if the validation loss does not (notably) improve anymore. Early stopping is both intuitive and effective to use, so it has become a standard addition for model training.
To better study the effect, we can now safely go back to models with many (too many?) parameters:
model = create_nn()
model.compile(optimizer='adam',
loss='mse',
metrics=[keras.metrics.RootMeanSquaredError()])
To apply early stopping during training it is easiest to use Keras EarlyStopping class.
This allows to define the condition of when to stop training. In our case we will say when the validation loss is lowest.
However, since we have seen quiet some fluctuation of the losses during training above we will also set patience=10 which means that the model will stop training if the validation loss has not gone down for 10 epochs.
from tensorflow.keras.callbacks import EarlyStopping
earlystopper = EarlyStopping(
monitor='val_loss',
patience=10,
verbose=1
)
history = model.fit(X_train, y_train,
batch_size = 32,
epochs = 200,
validation_data=(X_val, y_val),
callbacks=[earlystopper],
verbose = 2)
As before, we can plot the losses during training:
history_df = pd.DataFrame.from_dict(history.history)
sns.lineplot(data=history_df[['root_mean_squared_error', 'val_root_mean_squared_error']])
plt.xlabel("epochs")
plt.ylabel("RMSE")
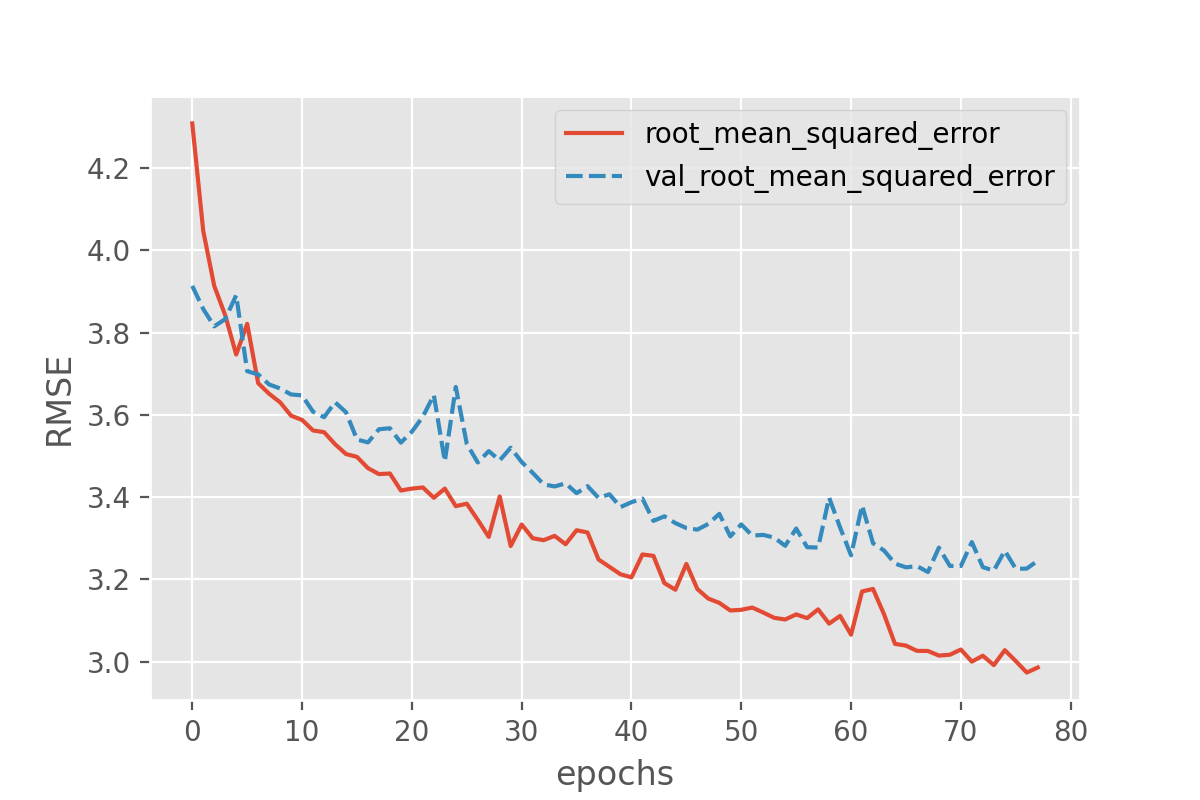
This still seems to reveal the onset of overfitting, but the training stops before the discrepancy between training and validation loss can grow further. Despite avoiding severe cases of overfitting, early stopping has the additional advantage that the number of training epochs will be regulated automatically. Instead of comparing training runs for different number of epochs, early stopping allows to simply set the number of epochs to a desired maximum value.
What might be a bit unintuitive is that the training runs might now end very rapidly. This might spark the question: have we really reached an optimum yet? And often the answer to this is “no”, which is why early stopping frequently is combined with other approaches to avoid overfitting. Overfitting means that a model (seemingly) performs better on seen data compared to unseen data. One then often also says that it does not “generalize” well. Techniques to avoid overfitting, or to improve model generalization, are termed regularization techniques and we will come back to this in episode 4.
BatchNorm: the “standard scaler” for deep learning
A very common step in classical machine learning pipelines is to scale the features, for instance by using sckit-learn’s StandardScaler.
This can in principle also be done for deep learning.
An alternative, more common approach, is to add BatchNormalization layers (documentation of the batch normalization layer) which will learn how to scale the input values.
Similar to dropout, batch normalization is available as a network layer in Keras and can be added to the network in a similar way.
It does not require any additional parameter setting.
from tensorflow.keras.layers import BatchNormalization
The BatchNormalization can be inserted as yet another layer into the architecture.
def create_nn():
# Input layer
inputs = keras.layers.Input(shape=(X_data.shape[1],), name='input')
# Dense layers
layers_dense = keras.layers.BatchNormalization()(inputs)
layers_dense = keras.layers.Dense(100, 'relu')(layers_dense)
layers_dense = keras.layers.Dense(50, 'relu')(layers_dense)
# Output layer
outputs = keras.layers.Dense(1)(layers_dense)
# Defining the model and compiling it
return keras.Model(inputs=inputs, outputs=outputs, name="model_batchnorm")
model = create_nn()
model.compile(loss='mse', optimizer='adam', metrics=[keras.metrics.RootMeanSquaredError()])
model.summary()
This new layer appears in the model summary as well.
Model: "model_batchnorm"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(None, 89)] 0
_________________________________________________________________
batch_normalization (BatchNo (None, 89) 356
_________________________________________________________________
dense (Dense) (None, 100) 9000
_________________________________________________________________
dense_1 (Dense) (None, 50) 5050
_________________________________________________________________
dense_2 (Dense) (None, 1) 51
=================================================================
Total params: 14,457
Trainable params: 14,279
Non-trainable params: 178
We can train the model again as follows:
history = model.fit(X_train, y_train,
batch_size = 32,
epochs = 1000,
validation_data=(X_val, y_val),
callbacks=[earlystopper],
verbose = 2)
history_df = pd.DataFrame.from_dict(history.history)
sns.lineplot(data=history_df[['root_mean_squared_error', 'val_root_mean_squared_error']])
plt.xlabel("epochs")
plt.ylabel("RMSE")
Batchnorm parameters
You may have noticed that the number of parameters of the Batchnorm layers corresponds to 4 parameters per input node. These are the moving mean, moving standard deviation, additional scaling factor (gamma) and offset factor (beta). There is a difference in behavior for Batchnorm between training and prediction time. During training time, the data is scaled with the mean and standard deviation of the batch. During prediction time, the moving mean and moving standard deviation of the training set is used instead. The additional parameters gamma and beta are introduced to allow for more flexibility in output values, and are used in both training and prediction,
Run on test set and compare to naive baseline
It seems that no matter what we add, the overall loss does not decrease much further (we at least avoided overfitting though!). Let’s again plot the results on the test set:
y_test_predicted = model.predict(X_test)
plt.figure(figsize=(5, 5), dpi=100)
plt.scatter(y_test_predicted, y_test, s=10, alpha=0.5)
plt.xlabel("predicted sunshine hours")
plt.ylabel("true sunshine hours")
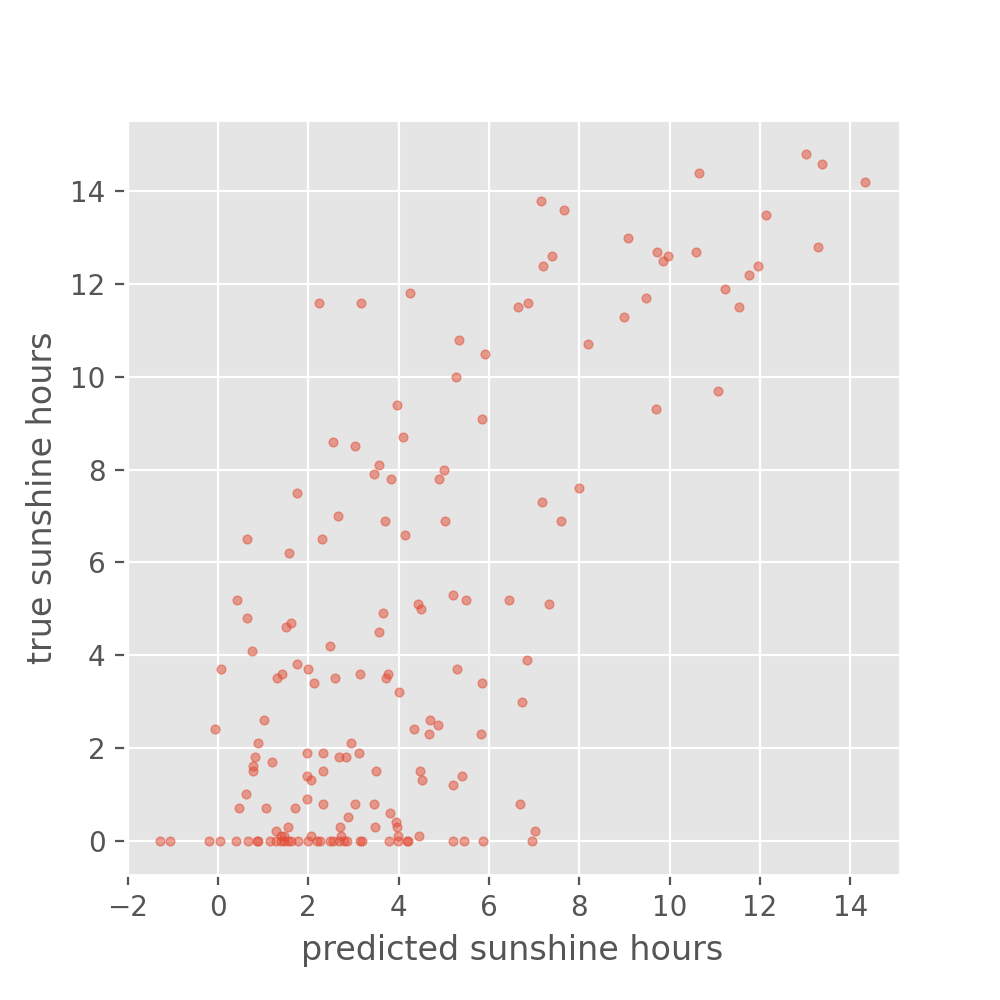
Well, the above is certainly not perfect. But how good or bad is this? Maybe not good enough to plan your picnic for tomorrow. But let’s better compare it to the naive baseline we created in the beginning. What would you say, did we improve on that?
Exercise: Simplify the model and add data
You may have been wondering why we are including weather observations from multiple cities to predict sunshine hours only in Basel. The weather is a complex phenomenon with correlations over large distances and time scales, but what happens if we limit ourselves to only one city?
- Since we will be reducing the number of features quite significantly, we should afford to include more data. Instead of using only 3 years, use 8 or 9 years!
- Remove all cities from the training data that are not for Basel. You can use something like:
cols = [c for c in X_data.columns if c[:5] == 'BASEL'] X_data = X_data[cols]- Now rerun the last model we defined which included the BatchNorm layer. Recreate the scatter plot comparing your prediction with the baseline prediction based on yesterday’s sunshine hours, and compute also the RMSE. Note that even though we will use many more observations than previously, the network should still train quickly because we reduce the number of features (columns). Is the prediction better compared to what we had before?
Solution
Use 9 years out of the total dataset. This means 3 times as many rows as we used previously, but by removing columns not containing “BASEL” we reduce the number of columns from 89 to 11.
nr_rows = 365*9 # data X_data = data.loc[:nr_rows].drop(columns=['DATE', 'MONTH']) # labels (sunshine hours the next day) y_data = data.loc[1:(nr_rows + 1)]["BASEL_sunshine"] # only use columns with 'BASEL' cols = [c for c in X_data.columns if c[:5] == 'BASEL'] X_data = X_data[cols]Do the train-test-validation split:
X_train, X_holdout, y_train, y_holdout = train_test_split(X_data, y_data, test_size=0.3, random_state=0) X_val, X_test, y_val, y_test = train_test_split(X_holdout, y_holdout, test_size=0.5, random_state=0)Function to create a network including the BatchNorm layer:
def create_nn(): # Input layer inputs = keras.layers.Input(shape=(X_data.shape[1],), name='input') # Dense layers layers_dense = keras.layers.BatchNormalization()(inputs) layers_dense = keras.layers.Dense(100, 'relu')(layers_dense) layers_dense = keras.layers.Dense(50, 'relu')(layers_dense) # Output layer outputs = keras.layers.Dense(1)(layers_dense) # Defining the model and compiling it return keras.Model(inputs=inputs, outputs=outputs, name="model_batchnorm")Create the network. Because we have reduced the number of input features the number of parameters in the network goes down from 14457 to 6137.
# create the network and view its summary model = create_nn() model.compile(loss='mse', optimizer='adam', metrics=[keras.metrics.RootMeanSquaredError()]) model.summary()Fit with early stopping and output showing performance on validation set:
history = model.fit(X_train, y_train, batch_size = 32, epochs = 1000, validation_data=(X_val, y_val), callbacks=[earlystopper], verbose = 2) # plot RMSE history_df = pd.DataFrame.from_dict(history.history) sns.lineplot(data=history_df[['root_mean_squared_error', 'val_root_mean_squared_error']]) plt.xlabel("epochs")Create a scatter plot to compare with true observations:
y_test_predicted = model.predict(X_test) plt.figure(figsize=(5, 5), dpi=100) plt.scatter(y_test_predicted, y_test, s=10, alpha=0.5) plt.xlabel("predicted sunshine hours") plt.ylabel("true sunshine hours")Compare the mean squared error with baseline prediction. It should be similar or even a little better than what we saw with the larger model!
from sklearn.metrics import mean_squared_error y_baseline_prediction = X_test['BASEL_sunshine'] rmse_nn = mean_squared_error(y_test, y_test_predicted, squared=False) rmse_baseline = mean_squared_error(y_test, y_baseline_prediction, squared=False) print('NN RMSE: {:.2f}, baseline RMSE: {:.2f}'.format(rmse_nn, rmse_baseline))
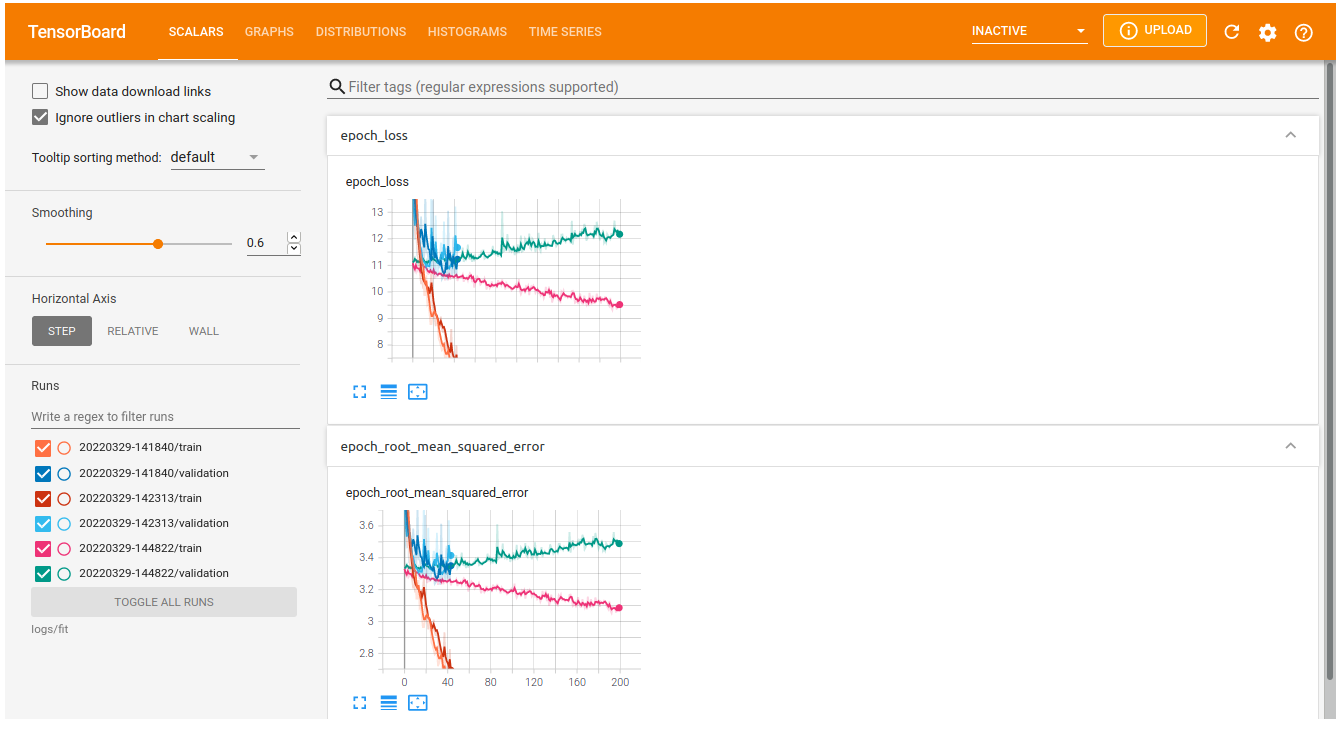
Tensorboard
If we run many different experiments with different architectures, it can be difficult to keep track of these different models or compare the achieved performance. We can use tensorboard, a framework that keeps track of our experiments and shows graphs like we plotted above. Tensorboard is included in our tensorflow installation by default. To use it, we first need to add a callback to our (compiled) model that saves the progress of training performance in a logs directory:
from tensorflow.keras.callbacks import TensorBoard import datetime log_dir = "logs/fit/" + datetime.datetime.now().strftime("%Y%m%d-%H%M%S") # You can adjust this to add a more meaningful model name tensorboard_callback = TensorBoard(log_dir=log_dir, histogram_freq=1) history = model.fit(X_train, y_train, batch_size = 32, epochs = 200, validation_data=(X_val, y_val), callbacks=[tensorboard_callback], verbose = 2)You can launch the tensorboard interface from a Jupyter notebook, showing all trained models:
%load_ext tensorboard %tensorboard --logdir logs/fitWhich will show an interface that looks something like this:
Outlook
Correctly predicting tomorrow’s sunshine hours is apparently not that simple. Our models get the general trends right, but still predictions vary quite a bit and can even be far off.
Open question: What could be next steps to further improve the model?
With unlimited options to modify the model architecture or to play with the training parameters, deep learning can trigger very extensive hunting for better and better results. Usually models are “well behaving” in the sense that small chances to the architectures also only result in small changes of the performance (if any). It is often tempting to hunt for some magical settings that will lead to much better results. But do those settings exist? Applying common sense is often a good first step to make a guess of how much better could results be. In the present case we might certainly not expect to be able to reliably predict sunshine hours for the next day with 5-10 minute precision. But how much better our model could be exactly, often remains difficult to answer.
- What changes to the model architecture might make sense to explore?
- Ignoring changes to the model architecture, what might notably improve the prediction quality?
Solution
This is on open question. And we don’t actually know how far one could push this sunshine hour prediction (try it out yourself if you like! We’re curious!). But there is a few things that might be worth exploring.
Regarding the model architecture:
- In the present case we do not see a magical silver bullet to suddenly boost the performance. But it might be worth testing if deeper networks do better (more layers).
Other changes that might impact the quality notably:
- The most obvious answer here would be: more data! Even this will not always work (e.g. if data is very noisy and uncorrelated, more data might not add much).
- Related to more data: use data augmentation. By creating realistic variations of the available data, the model might improve as well.
- More data can mean more data points (you can test it yourself by taking more than the 3 years we used here!)
- More data can also mean more features! What about adding the month?
- The labels we used here (sunshine hours) are highly biased, many days with no or nearly no sunshine but few with >10 hours. Techniques such as oversampling or undersampling might handle such biased labels better. Another alternative would be to not only look at data from one day, but use the data of a longer period such as a full week. This will turn the data into time series data which in turn might also make it worth to apply different model architectures…
Key Points
Separate training, validation, and test sets allows monitoring and evaluating your model.
Batchnormalization scales the data as part of the model.